Topics Map > Services > Research Computing and Support > CCAST
Highlights - Genomic Basis of Fetal Programming in Beef Cattle
Principal Investigators: Alison Ward, Carl Dahlen, Joel Caton, Lawrence Reynolds, and Wellison Diniz (Animal Sciences, North Dakota State University)
The long-term goals of this multidisciplinary research team are to understand the regulatory mechanisms that underlie developmental programming in beef cattle and ultimately to develop nutritional strategies to improve the efficiency of beef cattle production.
Maternal nutrition insults during critical windows of development impair offspring development and tissue programming [1]. Furthermore, several studies have highlighted the long-term adverse effects of maternal imbalances on offspring performance [2–4]. However, the genomic mechanisms driving fetal programming in response to maternal nutrition and other environmental factors are still to be elucidated.
To gain insights into the mechanisms involved with fetal programming, the team has developed a bovine pregnancy model where maternal, fetal, and placental tissues are collected at several time-points throughout gestation and birth [5]. Based on transcriptomics analysis (RNA-Seq), they assess how maternal nutrition leads to changes in gene expression profile in tissues such as the liver and muscle. Furthermore, they use co-expression network approaches (PCIT) to identify hub genes and key transcription factors (TFs) modulating gene expression; see Fig. 1.
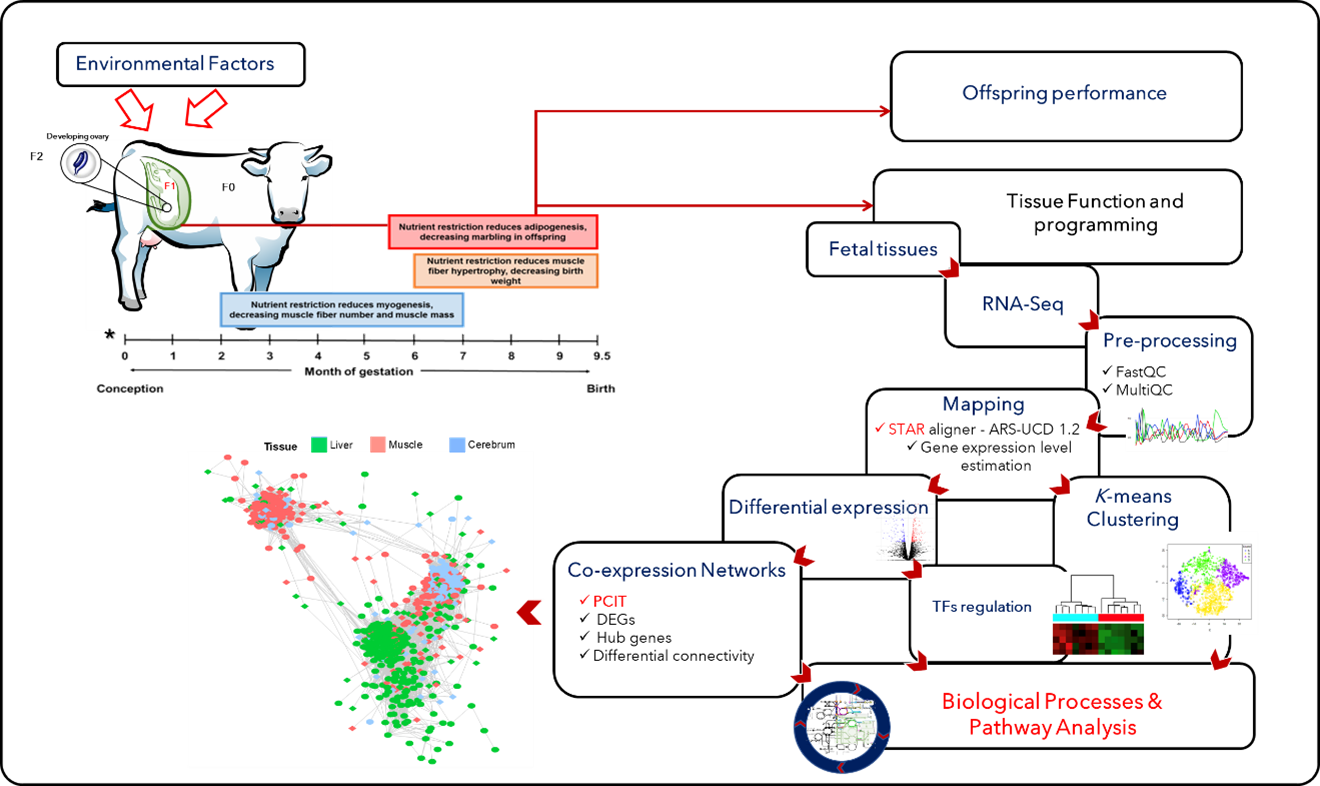
Figure 1: Bioinformatics approaches used to investigate the effects of maternal nutrition on gene transcription during fetal development and programming [1].
Due to the large amount of data and the increased computing power involved, these analyses are performed on high-performance computing (HPC) systems at CCAST where state-of-the-art bioinformatic tools are used for transcript mapping, gene annotation, co-expression network construction, and functional analyses.
Examples of their research outputs that were made possible by the use of CCAST resources include recent publications in Scientific Reports and Genes.
The projects are funded by the USDA-NIFA, ND State Board of Agricultural Research and Education, ND Agricultural Experimental Station, and Purina.
References
[1] Du, M.; Tong, J.; Zhao, J.; Underwood, K.R.; Zhu, M.; Ford, S.P.; Nathanielsz, P.W. Fetal programming of skeletal muscle development in ruminant animals. J. Anim. Sci. 2009, 88, E51.
[2] Caton, J.S.; Crouse, M.S.; McLean, K.J.; Dahlen, C.R.; Ward, A.K.; Cushman, R.A.; Grazul-Bilska, A.T.; Neville, B.W.; Borowicz, P.P.; Reynolds, L.P. Maternal periconceptual nutrition, early pregnancy, and developmental outcomes in beef cattle. J. Anim. Sci. 2020, 98, skaa358.
[3] Crouse, M.S.; Caton, J.S.; Cushman, R.A.; McLean, K.J.; Dahlen, C.R.; Borowicz, P.P.; Reynolds, L.P.; Ward, A.K. Moderate nutrient restriction of beef heifers alters expression of genes associated with tissue metabolism, accretion, and function in fetal liver, muscle, and cerebrum by day 50 of gestation. Transl. Anim. Sci. 2019, 3, 855.
[4] Peñagaricano, F.; Wang, X.; Rosa, G.J.M.; Radunz, A.E.; Khatib, H. Maternal nutrition induces gene expression changes in fetal muscle and adipose tissues in sheep. BMC Genomics 2014, 15, 1034.
[5] McLean, K.J.; Dahlen, C.R.; Borowicz, P.P.; Reynolds, L.P.; Crosswhite, M.R.; Neville, B.W.; Walden, S.D.; Caton, J.S. Technical note: A new surgical technique for ovariohysterectomy during early pregnancy in beef heifers. J. Anim. Sci. 2016, 94, 5089.
[6] Diniz, W.J.S.; Crouse, M.S.; Cushman, R.A.; McLean, K.J.; Caton, J.S.; Dahlen, C.R.; Reynolds, L.P.; Ward, A.K. Cerebrum, liver, and muscle regulatory networks uncover maternal nutrition effects in developmental programming of beef cattle during early pregnancy. Sci. Rep. 2021, 11, 2771.