Topics Map > Services > Research Computing and Support > CCAST
Highlights - Identification of Multi-Omic Marker Associations in Pancreatic Cancer
Principal Investigator: Rick Jansen (Public Health, North Dakota State University)
Performing epigenetic and epigenomic analysis is key to understanding biological and molecular tumor signatures in order to identify genomic markers to target for treatment or interventions. There have been many computationally-based genomic studies looking for single marker associations with pancreatic ductal adenocarcinoma (PDAC) using blood and/or tissue samples. However, relatively rarely have specific markers been validated or reproduced by multiple studies, because of several limitations including: limited power because of multiple statistical tests, high false positive rates, variability of platforms used, and sample quality and purity problems.
A research project in Dr. Jansen’s group, funded by NIH through the COBRE Center for Diagnostic and Therapeutic Strategies in Pancreatic Cancer at NDSU, uses publicly available PDAC datasets to implement several omics-based approaches to identify the PDAC-associated alterations across omic sets (expression, methylation, sequence, etc.) which are specifically associated with key identified core genes including KRAS, CDKN2A, TP53, and SMAD4 [1]. These genomic analyses are then validated on Mayo Clinic patient tumor samples to categorize the associations based on key demographic and clinical variables. The group is determining the important genomic marker sets for additional future research investigation.
Use existing whole genome datasets to create network associations with previously identified core genes:
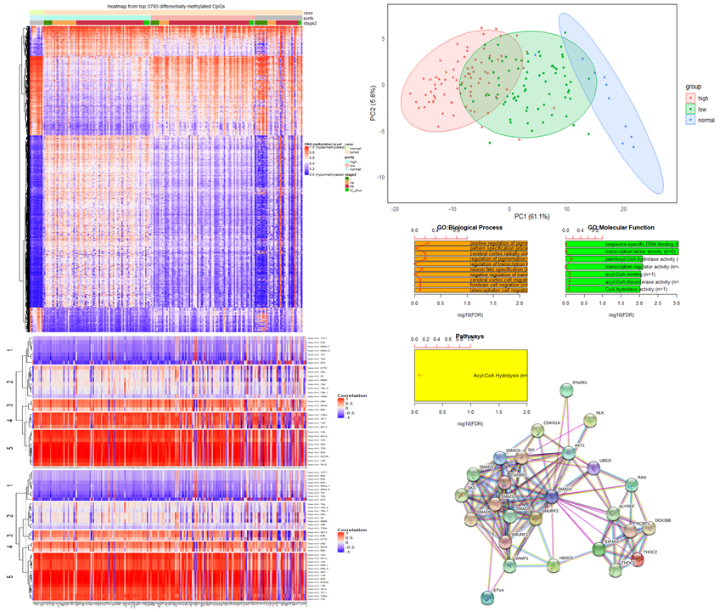
Figure 1: Muli-omics bioinformatics workflow using heatmaps, correlation, PCA, and GO enrichment analysis to identify genomic network associations important in PDAC.
This part of the project uses The Cancer Genome Atlas (TCGA) and the International Cancer Genome Consortium (ICGC) data. The focus is on combining gene sequence, gene expression, methylation, and clinical information using several developed methods (e.g., principal component and SMV) to identify genome-wide associations with PDAC compared with adjacent normal pancreas tissue [2,3]. Dr. Jansen’s group has constructed a network of important genomic alterations associated with each of the key core genes. The linking of multi-omics data and core genes give them a more complete picture as to the importance of alterations associated with each gene and the ultimate role across different subgroupings of PDAC.
Use PDAC patient tumor samples to categorize associations based on key demographic and clinical variables:

Figure 2: Visualization of deterministic causal models as presented by Rothman et al. [4].
The group uses the networks defined above to investigate correlation between those alterations within patient sample sets based on key demographic and clinical variables (e.g., are alterations more likely among older PDAC patients compared to younger or among ever smokers compared with never smokers). This part of the project allows them to present observed individual causal models and describe if they vary by key demographic/clinical variables in order to generate hypothesis about PDAC development and progression and develop causal models [4].
There is a great advantage in using publicly available data and exploratory-based methods to analyze multiple types of genomic markers. This step is relatively cost effective and efficient, as a way to improve power and resources in deeper analyses. High quality and complete data are essential along with a biological understanding of the importance of specific pathways in PDAC development. With this project, Dr. Jansen’s group will be linking tumor genomics and patient characteristics to be used in future research such as projects focused on understanding PDAC development and progression. Their framework has very high potential to be generalized to the analysis of other PDAC samples and other cancers or disease types. Additionally, the results can provide other researchers with a foundation around which to focus resources to create intervention or treatment targets.
The group relies on HPC resources at CCAST to improve the speed of analyses and efficiently download, store, and process these large genomic datasets.
References
[1] Jones, S.; Zhang, X.; Parsons, D.W.; Lin, J.C.-H.; Leary, R.J.; Angenendt, P.; Mankoo, P.; Carter, H.; Kamiyama, H.; Jimeno, A.; et al. Core signaling pathways in human pancreatic cancers revealed by global genomic analyses. Science 2008, 321, 1801–6.
[2] Kwon, M.-S.; Kim, Y.; Lee, S.; Namkung, J.; Yun, T.; Yi, S.G.; Han, S.; Kang, M.; Kim, S.W.; Jang, J.-Y.; et al. Integrative analysis of multi-omics data for identifying multi-markers for diagnosing pancreatic cancer. BMC Genomics 2015, 16 Suppl 9, S4.
[3] Rajamani, D.; Bhasin, M.K. Identification of key regulators of pancreatic cancer progression through multidimensional systems-level analysis. Genome Med. 2016, 8, 38.
[4] Rothman, K.J.; Greenland, S. Causation and causal inference in epidemiology. Am. J. Public Health 2005, 95.